ABSTRACT
Objective: Suicide is a priority health problem. Suicide assessment depends on imperfect clinician assessment with minimal ability to predict the risk of suicide. Machine learning/deep learning provides an opportunity to detect an individual at risk of suicide to a greater extent than clinician assessment. The present study aimed to use deep learning of structural magnetic resonance imaging (MRI) to create an algorithm for detecting suicidal ideation and suicidal attempts.
Methods: We recruited 4 groups comprising a total of 186 participants: 33 depressive patients with suicide attempt (SA), 41 depressive patients with suicidal ideation (SI), 54 depressive patients without suicidal thoughts (DP), and 58 healthy controls (HCs). The confirmation of depressive disorder, SA and SI was based on psychiatrists’ diagnosis and Mini-International Neuropsychiatric Interview (MINI) interviews. In the generalized q-sampling imaging (GQI) dataset, indices of generalized fractional anisotropy (GFA), the isotropic value of the orientation distribution function (ISO), and normalized quantitative anisotropy (NQA) were separately trained in convolutional neural network (CNN)–based deep learning and DenseNet models.
Results: From the results of 5-fold cross-validation, the best accuracies of the CNN classifier for predicting SA, SI, and DP against HCs were 0.916, 0.792, and 0.589, respectively. In SA-ISO, DenseNet outperformed the simple CNNs with a best accuracy from 5-fold cross-validation of 0.937. In SA-NQA, the best accuracy was 0.915.
Conclusions: The results showed that a deep learning method based on structural MRI can effectively detect individuals at different levels of suicide risk, from depression to suicidal ideation and attempted suicide. Further studies from different populations, larger sample sizes, and prospective follow-up studies are warranted to confirm the utility of deep learning methods for suicide prevention and intervention.
J Clin Psychiatry 2021;82(2):19m13225
To cite: Chen VC-H, Wong F-T, Tsai Y-H, et al. Convolutional neural network–based deep learning model for predicting differential suicidality in depressive patients using brain generalized q-sampling imaging. J Clin Psychiatry. 2021;82(2):19m13225.
To share: https://doi.org/10.4088/JCP.19m13225
© Copyright 2021 Physicians Postgraduate Press, Inc.
aSchool of Medicine, Chang Gung University, Taoyuan, Taiwan
bDepartment of Psychiatry, Chang Gung Memorial Hospital, Chiayi, Taiwan
cDepartment of Medical Imaging and Radiological Sciences, Bachelor Program in Artificial Intelligence, Chang Gung University, Taoyuan, Taiwan
dDepartment of Diagnostic Radiology, Chang Gung Memorial Hospital, Chiayi, Taiwan
eDepartment of Counseling and Clinical Psychology, Columbia University, New York City, New York
fMood Disorder Psychopharmacology Unit, University Health Network, Department of Psychiatry, University of Toronto, Ontario, Canada
gInstitute of Medical Science, University of Toronto, Toronto, Ontario, Canada
hDepartments of Psychiatry and Pharmacology, University of Toronto, Toronto, Ontario, Canada
iMedical Imaging Research Center, Institute for Radiological Research, Chang Gung University and Chang Gung Memorial Hospital at Linkou, Taoyuan, Taiwan
*Corresponding author: Jun-Cheng Weng, PhD, Department of Medical Imaging and Radiological Sciences, Chang Gung University, No. 259, Wenhua 1st Rd, Guishan Dist., Taoyuan City 33302, Taiwan ([email protected]).
Suicide is an important and serious public health problem worldwide and one of the most severe outcomes of health. In 2012, approximately 800,000 people died of self-inflicted injuries.1 Suicide is conceptualized as a process of phenotypically overlapping notions of ideations and behaviors from mild to more severe forms of suicidality, most often including the following stages: suicidal ideation, suicide plan, attempted suicide, and completed suicide.2
Assessment of suicide risk is one of the most challenging issues in the care of patients. The evaluation of suicide risk mostly depends on an evaluation by clinicians based on phenomenological and family history data, which provide suboptimal prediction. According to a recent systematic review3 regarding suicide prediction models, many common risk factors for suicide are not specific to individuals who engage in suicide attempts, resulting in a difficulty of diagnostic accuracy. Patient denial of suicide risk can easily lead to reduced vigilance during suicide assessment.2 The limited ability of current suicide assessments is illustrated by the findings of a recent meta-analysis4 regarding suicide rates after discharge from psychiatric facilities, wherein it was observed that the suicide rate was highest within 3 months after discharge.
Several efforts have been made to address deficiencies in current risk assessment techniques, including investigations of objective evaluation tools such as neuroimaging techniques. Extant neuroimaging studies have suggested that among individuals who are suicidal, the frontotemporal network, primarily representing reductions of gray and white matter volumes in the prefrontal cortex, anterior cingulate, and superior temporal gyrus, is involved.5 The majority of magnetic resonance imaging (MRI) studies based on traditional statistics report group differences between persons with suicide risk and those without suicide risk. Emerging analysis techniques, such as machine learning or deep learning, have shed light on this issue to create algorithms to identify which individual is at risk.6 To date, only one recent study7 has been reported on the use of a machine-learning algorithm based on functional MRI neural signatures of death and life-related concepts for detecting young individuals with suicidal ideation with 91% accuracy. There has been no related study exploring whether deep learning methods based on magnetic resonance (MR) images can be an identification tool for suicide.
The present study aimed to investigate whether MRI-measured structural changes can assist risk stratification among those with different levels of suicide risk—individuals without depression, depressed patients, patients with suicidal ideation, and patients who have attempted suicide—based on a deep learning method.
METHODS
Participants
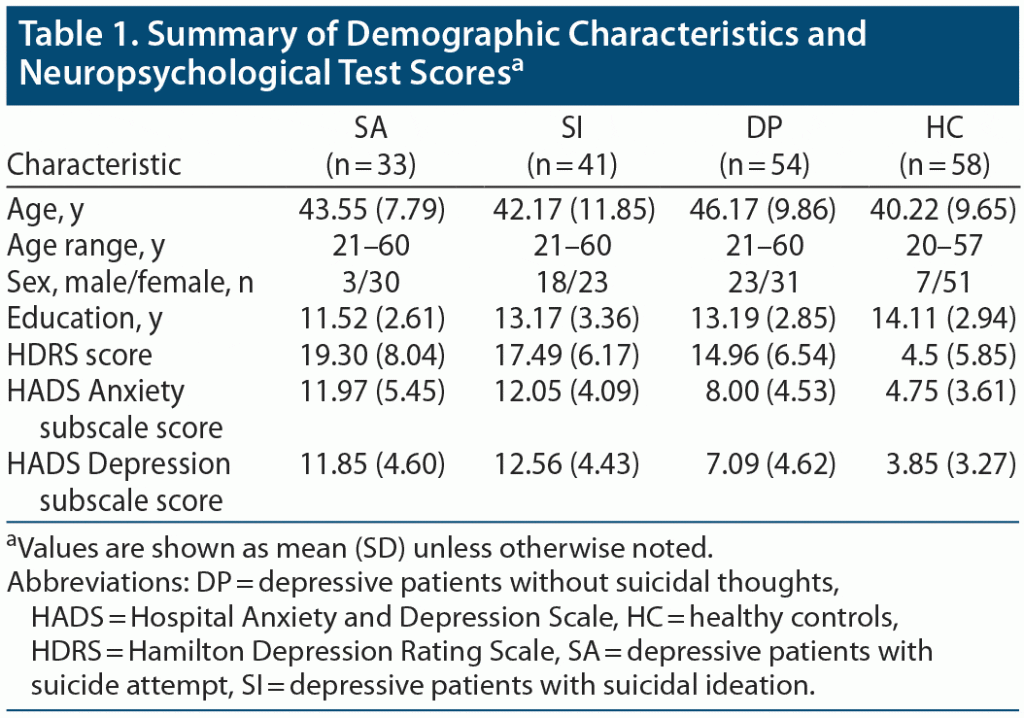
We recruited 4 groups of 186 participants: 33 major depressive disorder patients (MDD) who have made at least 1 suicide attempt (SA) (aged 21–60 years, mean
[SD] = 43.55 [7.79] years), 41 MDD patients with suicidal ideation (SI) (aged 21–60 years, mean [SD] = 42.17 [11.85] years), 54 patients with MDD but without suicidal thoughts (DP) (aged 21–60 years, mean [SD] = 46.17 [9.86] years), and 58 healthy controls (HCs) (aged 20–57 years, mean [SD] = 40.22 [9.65] years). The confirmation of MDD, suicidal ideation, and attempted suicide was primarily based on psychiatrists’ diagnosis and Mini-International Neuropsychiatric Interview (MINI)8 interviews carried out by trained research nurses. The final confirmation was carried out by the principal investigator (V.C.-H.C.) according to all available information. All participants were at least 20 years of age and right-handed. The participants were recruited via the outpatient clinic of the Department of Psychiatry at Chiayi Chang Gung Memorial Hospital and recruitment advertisements. Exclusion criteria for all participants were any eye diseases (eg, cataract and glaucoma), history of another primary mental disorder (eg, schizophrenia or bipolar disorder), alcohol or illicit substance use disorder during the past year, any neurologic illnesses, and metallic implants or other contraindications for MRI. The study was approved by the Institutional Review Board of Chang Gung Memorial Hospital, Chiayi, Taiwan (No. 104–0838B, 104–9337B, 201602027B0). All participants joined the study after providing written informed consent, and all research was performed in accordance with relevant guidelines and regulations. The datasets generated and/or analyzed during the current study are not publicly available owing to data privacy policy at our facility, but are available from the corresponding author on reasonable request.
Diffusion MRI Data Acquisition
All participants were scanned using a 3T MRI system (Verio; SIEMENS, Germany) with a single-shot, diffusion-weighted spin-echo echo-planar imaging sequence. Diffusion images were obtained with repetition time/echo time (TR/TE) = 8,943/115 ms, field of view (FOV) = 250 × 250 mm2, matrix = 128 × 128, slices = 35, in-plane resolution = 2 × 2 mm2, slice thickness = 4 mm, signal average = 1, and 192 noncollinear diffusion weighting gradient direction with b values = 1,000, 1,500, and 2,000 s/mm2 and 1 null image without diffusion weighting (b value = 0 s/mm2).
Generalized q-Sampling Imaging Analysis
Based on the Fourier transform between the diffusion MR signals and the diffusion displacement, a new relationship can be deduced by directly estimating the spin distribution function (SDF) from the diffusion MR signals. This relationship leads to a new reconstructed method called generalized q-sampling imaging (GQI). GQI can provide directional and quantitative information about crossing fibers. GQI is a model-free method that quantifies the density of water, which diffuses in different orientations. Model-free methods estimate the empirical distribution of water diffusion, and there is no hypothesis on the distribution. The SDF is the density of the diffusing water in different directions and is a kind of diffusion orientation distribution function (ODF). GQI provides information on the relation between the diffusion signals of water and the SDF. Studies have shown that GQI has good sensitivity and specificity for white matter properties and pathology.9
In GQI analysis, FMRIB Software Library (FSL) eddy current correction (FMRIB; Oxford, UK) was performed; subsequently, the corrected diffusion images were registered to the b0 (null) image in native diffusion space using a linear transformation. The registered images were mapped to the standard T2 template after an affine transformation with 12 degrees of freedom and nonlinear warps using Statistical Parametric Mapping software (SPM8; The Wellcome Centre for Human Neuroimaging, UCL Queen Square Institute of Neurology; London, UK). After the preprocessing procedure, GQI index mapping, including generalized fractional anisotropy (GFA), normalized quantitative anisotropy (NQA), and the isotropic value of the orientation distribution function (ISO), were reconstructed from multishell diffusion data using DSI Studio (National Taiwan University; Taipei, Taiwan). The GFA results indicate the anisotropy of the water diffusion in microstructures and is calculated from an orientation ODF. GFA is defined as the standard deviation divided by the root mean square of the ODF, indicating a measurement of the anisotropy. Quantitative anisotropy (QA) is calculated from the peak orientations on a spin distribution function and is defined for each fiber orientation. NQA is the normalized QA. ISO is the minimum distribution value of the ODF, and thus ISO represents the background isotropic diffusion.9
Convolutional Neural Network–Based Deep Learning
The GQI dataset included 3 derived indices: GFA, ISO, and NQA; our deep learning models were separately trained for each GQI index. The training and testing datasets were 2-dimensional images generated from the 3-dimensional volumes of each subject. The images were composed of slices in 3 planes, ie, axial, sagittal, and coronal views. Originally, the array shape of the 3-dimensional GQI volume was 91-by-109-by-91 voxels; to ensure that all the slices followed the same 2-dimensional image shape, each image slice was zero-padded to conform to a shape of 109-by-109 pixels. Hence, each volume can be sliced into 291 images. The dataset was divided into 5 groups for evaluating the results of 5-fold cross-validation. Therefore, the prediction results were the mean score across 5 independent models based on each of the corresponding 5-fold holdout samples, which were novel to our classifier. The training images were augmented by rotating the image twice. In the first step, each training image was separately randomly rotated between ± 25 degrees. Concatenating the original training images and the output of the first rotation, the new training images were made. In the second step, the same augmentation procedure was applied to the new training images with rotation angle between ± 40 degrees. Each rotating augmentation doubled the amount of data. In each of our binary classification tasks, images from the SA and DP groups separately served as our target group, while images from the HC group were our baseline group.
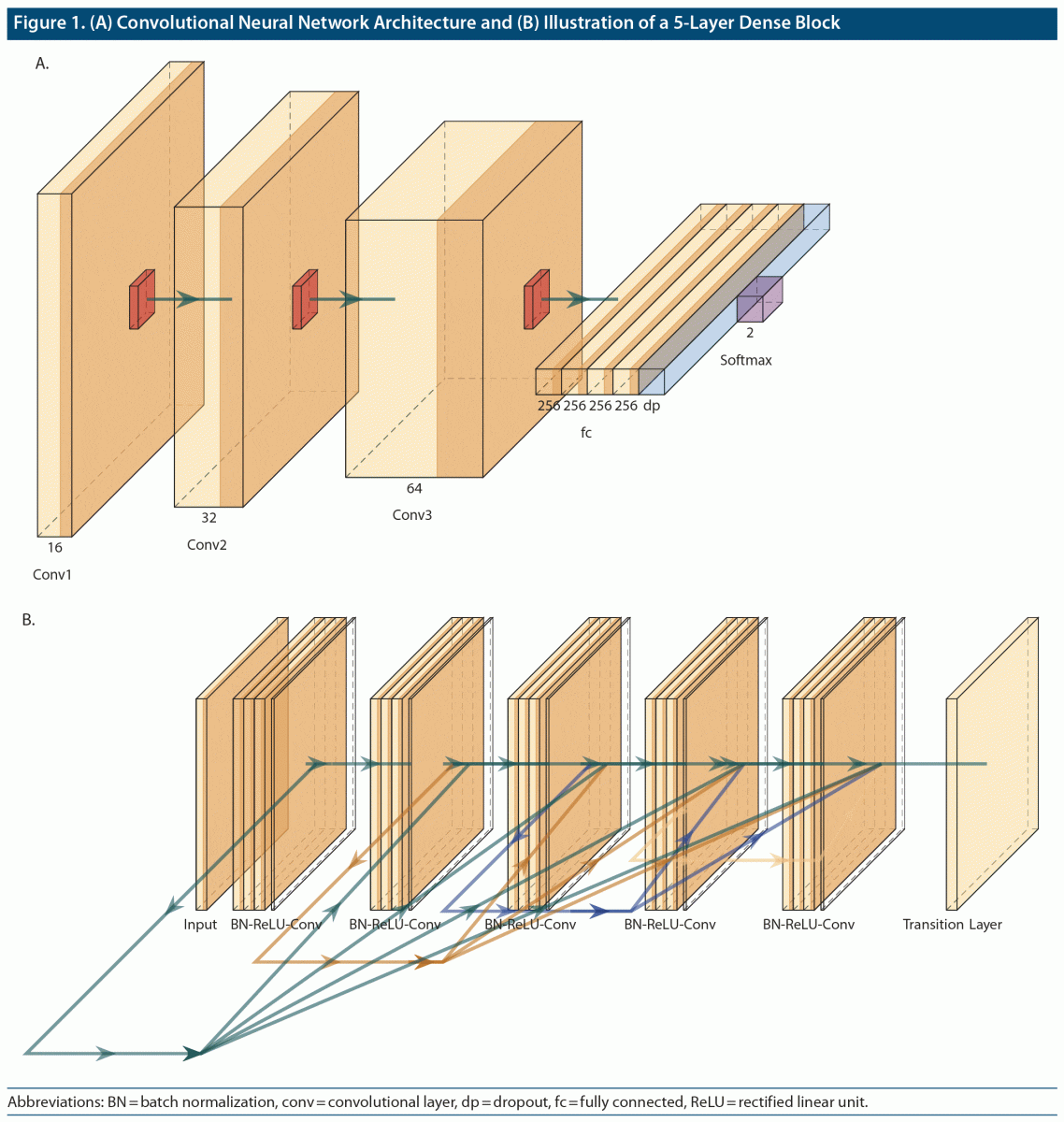
The architecture of the employed convolutional neural networks (CNNs) contained 3 convolution layers (Figure 1A), each of which (kernel size: 3-by-3 pixels, stride: 1-by-1 pixel) was followed by a max pooling layer (kernel size: 2-by-2 pixels). The number of filter maps in the 3 convolutional layers was 16, 32, and 64, sequentially. After flattening the last max pooling layer, there were four 256-unit fully connected layers and a dropout layer with a dropout rate of 0.3. A rectified linear unit (ReLU) was implemented as an activation function in each of the aforementioned convolutional layers. The classification results were obtained through a softmax layer. Adam optimizers were employed in the training process, and parameters were updated with loss scores weighted by the sample size in each corresponding group. Cross-entropy was applied as our loss function. The model was trained with a batch size of 32 and 30 epochs of training. The deep learning models were implemented in the TensorFlow framework (www.tensorflow.org).10
DenseNet
Furthermore, to test whether deeper CNNs could extract more predictive feature representations from our datasets, we also adopted a deeper but efficient CNN architecture—Dense Convolutional Network (DenseNet).11 The advantage of DenseNet is that it enables feature reuse and implicit deep supervision by implementations that allow each layer to receive feature inputs from all preceding layers to transmit its own feature maps to all subsequent layers. Figure 1B shows a dense block with 5 layers. In addition, a transfer learning scheme was adopted by initiating model weights with pretrained weights on ImageNet (www.image-net.org). To better fine-tune the model, we appended 2 fully connected layers (with 1,000 and 500 units, sequentially) at the end of the model architecture, which was implemented in the TensorFlow framework. The model was trained with a batch size of 32 and 30 epochs of training.
RESULTS
Participants
Table 1 shows the demographic characteristics of the participants. There were significant differences (P < .05) in age, sex, and years of education among the 3 groups per analysis of covariance. Therefore, age, sex, and years of education were used as covariates for subsequent analyses.
CNN-Based Deep Learning
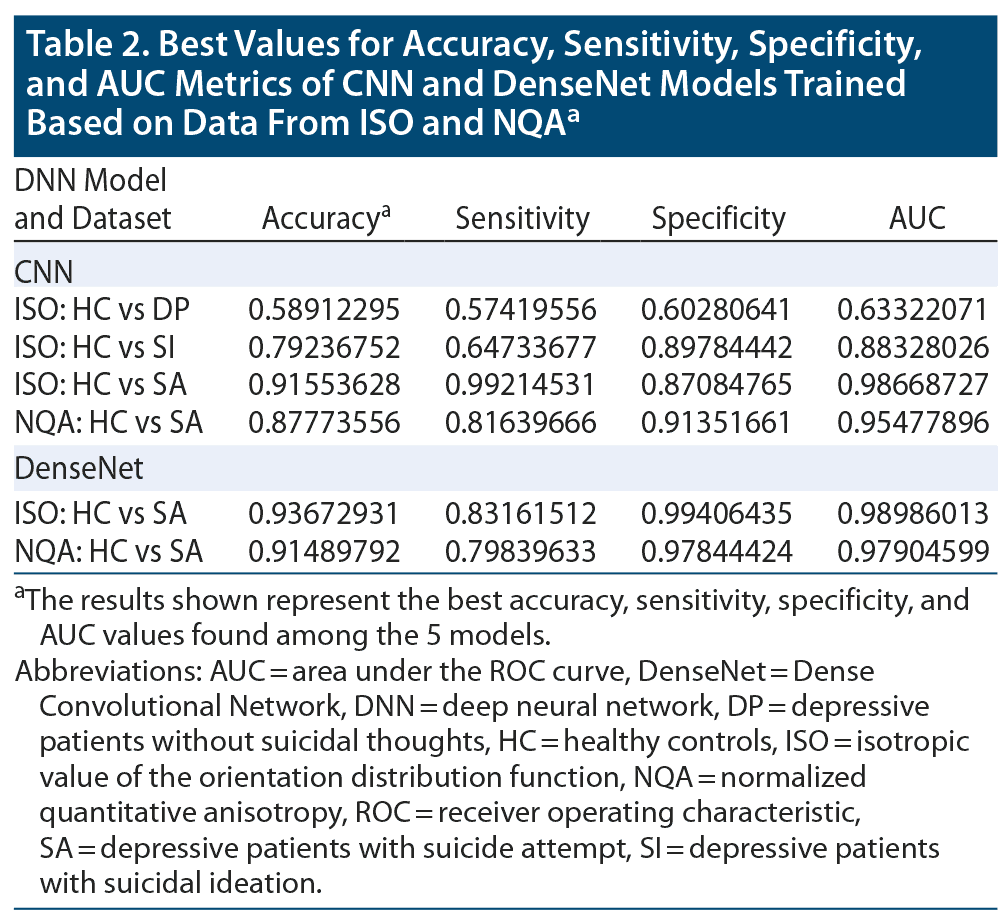
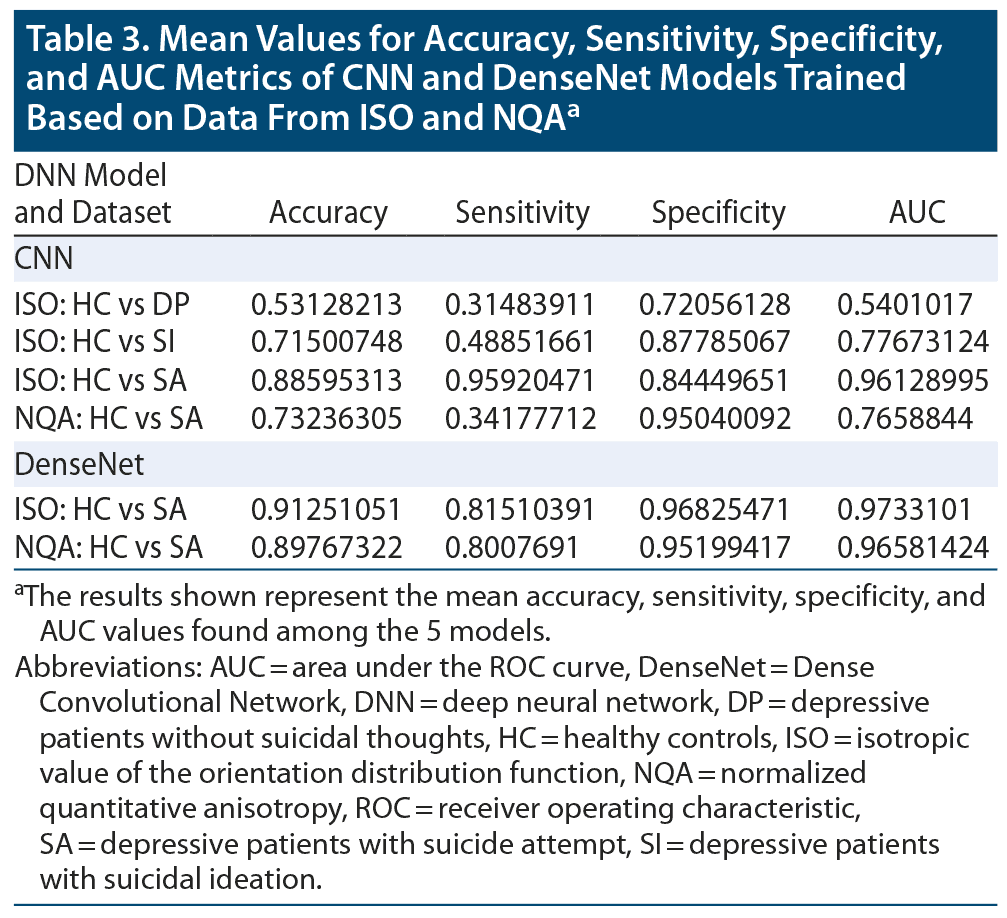
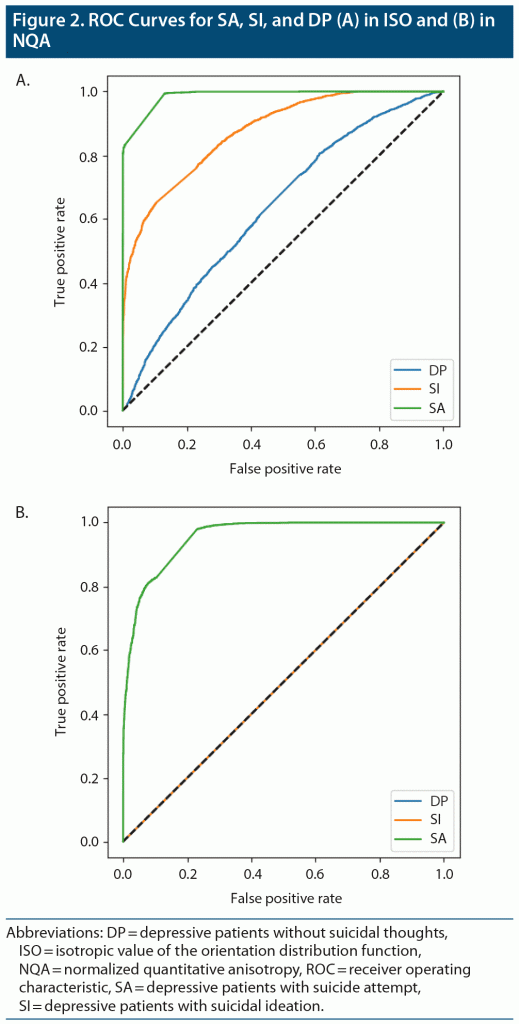
By averaging the results of the 5-fold cross-validation, the accuracies of our CNN classifier for predicting SA, SI, and DP against HC were 0.886 (0.916 was the best accuracy among the 5 models in SA), 0.715 (0.792 was the best accuracy among the 5 models in SI), and 0.531 (0.589 was the best accuracy among the 5 models in DP) in ISO. In NQA, the accuracy was 0.732 (0.878 was the best accuracy among the 5 models) in SA vs HC. Except for the aforementioned models, models trained based on NQA and with the SI and DP groups could not extract predictive features. The mean sensitivity (SEN), specificity (SPE), and area under the receiver operator characteristic curve (AUC) in SA-ISO were 0.959, 0.844, and 0.961, respectively. In SA-NQA, the mean SEN, SPE, and AUC were 0.341, 0.950, and 0.766, respectively. Other metrics of the best and mean results are listed in Table 2 and Table 3, respectively. Figure 2 illustrates the ROC curves predicted by the best model incorporating SA, SI, and DP in ISO and SA, SI, and DP in NQA.
DenseNet
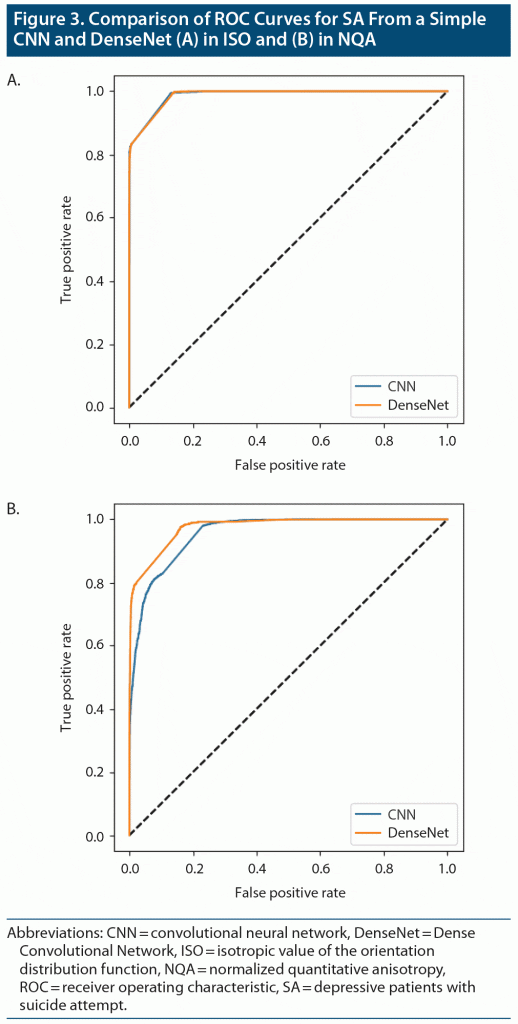
The results obtained from DenseNet were polarized in our applied datasets. In SA-ISO, DenseNet outperformed the simple CNNs with a mean accuracy from the 5-fold cross-validation of 0.913 (0.937 was the best accuracy among the 5 models). In SA-NQA, only 2 of the 5 models were able to learn meaningful representations. The mean accuracy of the 2 models was 0.898 (0.915 was the best accuracy among the 5 models). Other metrics of the best and mean results are summarized in Table 2 and Table 3, respectively. Figure 3 illustrates the comparisons of the ROC curves for SA in ISO and for SA in NQA from our simple CNN with those of DenseNet.
DISCUSSION
To the best of our knowledge, this study is the first to use deep learning methods based on structural MRI data to predict suicidality. The results suggested that, compared with other brain imaging indices, GQI-ISO possessed relatively more distinctive features for our implemented model, ie, a CNN for predicting attempted suicide. In addition, the accuracies among SA, SI, and DP in ISO suggested a gradient pattern, which is in accordance with the gradient pattern in terms of distinctive neural signatures observed among the aforementioned groups. In the results of the 5-fold cross-validation, the best accuracy scores of our CNN classifier for predicting SA, SI, and DP against HC were 0.916, 0.792, and 0.589, respectively. In SA-ISO, DenseNet outperformed the simple CNNs with a best accuracy from 5-fold cross-validation of 0.937. In SA-NQA, the best accuracy was 0.915.
To our knowledge, our study’s approach is the first to adopt the end-to-end CNN architecture without theoretically biased, human hand–crafted features, hence harnessing the power of the deep convolutional neuronal network. The performance of our classifier was better than that of a recently reported machine learning algorithm12 that was based on several steps that employ human hand–crafted feature selection procedure. It should also be noted that the metrics from the neural imaging database used by Gosnell et al12 are volume, cortical surface and thickness, and resting-state functional connectivity, while our best model was trained on GQI-reconstructed ISO.
Just et al7 employed a Gaussian naive Bayes classifier, which identified concepts and brain locations that were most predictive. Their approach is based on neurosemantic signatures, which rely on task-based functional MRI acquisition. Although it is of scientific value to explore the neural fingerprints of suicidal thoughts and behavior, the acquired data are limited in terms of clinical implementation due to cost and issues of accessibility.
Interestingly, our classifier learned the best feature representations from the isotropic diffusion component of the spin distribution value, ie, ISO.9 In terms of fiber tracking, subtracting ISO from the resolved fiber orientation has been claimed to reconstruct a more stable resolved fiber tracking result. Detailed information is defined in the equation from Yeh et al9: QA(â) = Z0(ψQ(â) – I(ψQ)), for which QA (in the resolved fiber orientation â) is calculated by the SDF value at the resolved orientation (ψQ(â)) minus the background isotropic diffusion component (I(ψQ)). Z0 is a scaling constant that scales free water diffusion to 1. Therefore, further investigations regarding the biophysiologic properties of ISO are needed to more fully inform our findings. QA is a more robust index than GFA, which is susceptible to several biological factors.13 Notwithstanding the fact that the GFA was a metric reconstructed by GQI, our current CNN classifier still did not learn predictive features from it.
Informed by a visual neuroscience model of the primate cortex, a series of filters of different sizes were implemented to extract complex and invariant feature representations that are highly informative for classification tasks in computer vision.14 Grid search is a well-known algorithm for tuning hyper-parameters in the machine leaning community, and tuning hyper-parameters is an important step of algorithm development.15 We found that homogeneous filters of scale 3-by-3 through our convolutional neural network yielded a more stable and better results for our GQI input images of shape 109-by-109 pixels. A filter size of 3-by-3 corresponds to the filter size used in mainstream implementations of the convolutional neural network; therefore, some detailed classifier design for our neural imaging task may still rely on the transferred knowledge from the research field of computer vision.
When a transfer learning scheme was applied, we obtained better model performance. However, this performance increment was not obvious in SA-ISO, from which our simple CNNs learned the best discriminative ability, suggesting an almost saturated level for CNNs to extract predictive representations. DenseNet architecture inherits the properties of identity mappings, deep supervision, and diversified depth; it is designed to retain compact internal representations and reduced feature redundancy, ideally making it a good feature extractor.11 Indeed, the revealed distinct performance increment in the SA-NQA dataset supports this claim. Nevertheless, the DenseNet performance was not stable across the models trained using the 5-fold cross-validation, which may be attributed to the sufficiency of the amount of data.
In addition, CNNs perform very well in a broad area on the computer vision area, especially in tasks related to natural images. Translational invariance is an important property of the data that CNNs are capable of detecting. An efficient approach to exploiting structure in the data may be the incorporation of structuring clusters of correlated features. Indeed, a recent study16 employing a stochastic regularizer based on feature clustering and averaging demonstrated better results than CNNs, especially in experimental conditions with small sample sizes and a low signal-to-noise ratio. Tractography datasets, as we applied here, are enriched with highly structural information. Whether it would be more informative to leverage the spatial patterns of the brain in our tractography dataset by structured regularization with the Recursive Nearest Agglomeration clustering scheme employed in the aforementioned study requires verification.17
In the broader perspective of a diathesis-stress model,18 our CNN model could act as a good candidate of predisposition to predict the risk of suicide attempts based on information from patients’ brain structures. With the aid of other stress-related societal factors, which potentially fluctuate across different phases of patients’ lifetimes and therefore are monitored from time to time in a clinical setting, critical periods should be identified in patients picked by our CNN model. Hence, we hope that the combined information flow would provide more precise predictions and help clinicians focus their limited resources on imminent high-risk patients and within a particular time period, such as those suffering suicidal crises or in an intensive outpatient program.
The current study may be limited by several factors. First, the generation ability of our models may be limited because samples were drawn from a single site and the same ethnic population. However, with the initial success to prove the feasibility of the end-to-end CNN architecture, inclusion of samples across sites and different ethnic population is worth investigating in future studies. Second, the interpretability of our deep learning model may be limited by the black-box nature of our classifiers. Future studies should aim to design variants of architecture to visualize important brain source patterns and to realize the goal of interpretable machine learning. Finally, the best performing model was predicting SA versus HC, but the results may be confounded by structural damage caused by the suicide attempt itself, such as gas intoxication. Further studies are warranted to repeat the analysis method in those with suicide attempt but whose suicide methods have no prominent direct brain damage, such as self-cutting.
CONCLUSION
It is difficult to predict suicide with phenomenology only, and there is a desire to inform risk by using image markers. Our results showed that a deep learning method based on structural MRI can effectively detect individuals at different levels of suicide risk, from depression to suicidal ideation and attempted suicide. Further studies from different populations and larger sample sizes and prospective follow-up studies are warranted to confirm the utility of deep learning methods for suicide prevention and intervention.
Submitted: December 25, 2019; accepted September 8, 2020.
Published online: February 23, 2021.
Potential conflicts of interest: All authors declare that they have no conflict of interest.
Funding/support: This study was supported by the research programs MOST105-2314-B-182-028, MOST106-2314-B-182-040-MY3, and MOST109-2314-B-182-047-MY3, which were sponsored by the Ministry of Science and Technology, Taipei, Taiwan.
Role of the sponsor: The funders had no role in the conduct of the study nor the collection, analysis, and interpretation of data.
Clinical Points
- Although suicidality is difficult to predict based on common risk factors for suicide, this study showed that a deep learning method based on structural magnetic imaging can effectively detect individuals at different levels of suicide risk, from depression to suicidal ideation and attempted suicide.
- The automated algorithm as a screening test can help doctors focus their limited resources on suicidal people who are emergently in need of help and make medical resources accessible to more people.
Editor’s Note: We encourage authors to submit papers for consideration as a part of our Focus on Suicide section. Please contact Philippe Courtet, MD, PhD, at [email protected].
References (18)

- Preventing suicide: A global imperative. World Health Organization. 2014. https://www.who.int/mental_health/suicide-prevention/world_report_2014/en/
- Sveticic J, De Leo D. The hypothesis of a continuum in suicidality: a discussion on its validity and practical implications. Ment Illn. 2012;4(2):e15. PubMed NLM
- Belsher BE, Smolenski DJ, Pruitt LD, et al. Prediction models for suicide attempts and deaths: a systematic review and simulation. JAMA Psychiatry. 2019;76(6):642–651. PubMed CrossRef NLM
- Chung DT, Ryan CJ, Hadzi-Pavlovic D, et al. Suicide rates after discharge from psychiatric facilities: a systematic review and meta-analysis. JAMA Psychiatry. 2017;74(7):694–702. PubMed CrossRef NLM
- Bani-Fatemi A, Tasmim S, Graff-Guerrero A, et al. Structural and functional alterations of the suicidal brain: an updated review of neuroimaging studies. Psychiatry Res Neuroimaging. 2018;278:77–91. PubMed CrossRef NLM
- Lee Y, Ragguett RM, Mansur RB, et al. Applications of machine learning algorithms to predict therapeutic outcomes in depression: a meta-analysis and systematic review. J Affect Disord. 2018;241:519–532. PubMed CrossRef NLM
- Just MA, Pan L, Cherkassky VL, et al. Machine learning of neural representations of suicide and emotion concepts identifies suicidal youth. Nat Hum Behav. 2017;1(12):911–919. PubMed CrossRef NLM
- Sheehan DV, Lecrubier Y, Sheehan KH, et al. The Mini-International Neuropsychiatric Interview (MINI): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59(suppl 20):22–33, quiz 34–57. PubMed NLM
- Yeh FC, Wedeen VJ, Tseng WY. Generalized q-sampling imaging. IEEE Trans Med Imaging. 2010;29(9):1626–1635. PubMed CrossRef NLM
- Abadi M, Agarwal A, Barham P, et al. TensorFlow: large-scale machine learning on heterogeneous distributed systems. Preliminary White Paper: November 9, 2015. https://arxiv.org/pdf/1603.04467.pdf.
- Huang G, Liu Z, van der Maaten L, et al. Densely Connected Convolutional Networks. Cornell University. 2017. http://arxivorg/abs/1608.06993
- Gosnell SN, Fowler JC, Salas R. Classifying suicidal behavior with resting-state functional connectivity and structural neuroimaging. Acta Psychiatr Scand. 2019;140(1):20–29. PubMed CrossRef NLM
- Yeh FC, Verstynen TD, Wang Y, et al. Deterministic diffusion fiber tracking improved by quantitative anisotropy. PLoS One. 2013;8(11):e80713. PubMed CrossRef NLM
- Serre T, Wolf L, Bileschi S, et al. Robust object recognition with cortex-like mechanisms. IEEE Trans Pattern Anal Mach Intell. 2007;29(3):411–426. PubMed CrossRef NLM
- Bergstra J, Bengio Y. Random search for hyper-parameter optimization. J Mach Learn Res. 2012;13:281–305.
- Aydore S, Thirion B, Varoquaux G. Feature grouping as a stochastic regularizer for high-dimensional structured data. Cornell University. 2019. https://arxiv.org/abs/1807.11718
- Hoyos-Idrobo A, Varoquaux G, Kahn J, et al. Recursive Nearest Agglomeration (ReNA): fast clustering for approximation of structured signals. IEEE Trans Pattern Anal Mach Intell. 2019;41(3):669–681. PubMed CrossRef NLM
- Rubinstein DH. A stress-diathesis theory of suicide. Suicide Life Threat Behav. 1986;16(2):182–197. PubMed CrossRef NLM
Please sign in or purchase this PDF for $40.
Save
Cite